Dr. Jinchao Li | 李金超

Our laboratory is dedicated to the study of DNA damage and the alteration of epigenetic modifications. We use Arabidopsis thaliana and soybean as model organisms and integrate multidisciplinary approaches from genetics, cell biology, biochemistry, and biophysics. Combined with high-throughput sequencing technologies and bioinformatics analysis methods, we systematically dissect the mechanisms of DNA damage perception, repair, and signaling in plants, and delve into the molecular and genetic basis of epigenetic modification variations caused by DNA damage.
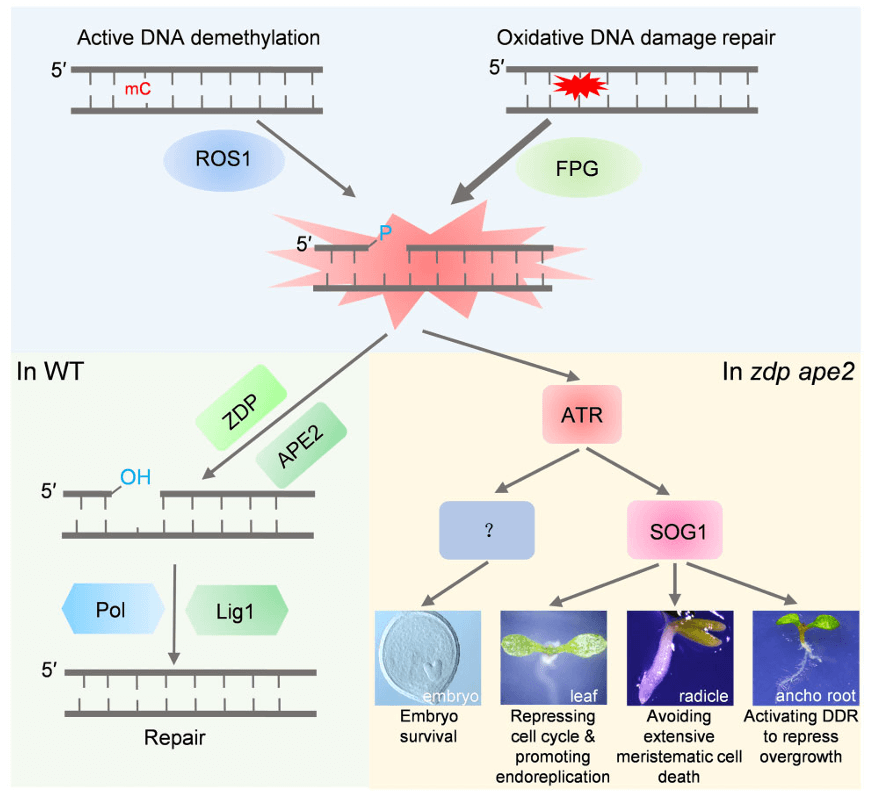
A Working Model of DNA 3′-Block-Triggered DNA Damage Response (DDR).
FPG-mediated base excision repair (BER) and ROS1-mediated DNA demethylation generate repair intermediates with a 3′-phosphate (3′-P) on the nicked strand. In wild-type (WT) plants, ZDP and APE2 convert this 3′-P to 3′-hydroxyl (3′-OH), allowing gap filling by DNA polymerase and sealing by DNA ligase 1. However, in zdp ape2 double mutants, unrepaired 3′-P molecules accumulate, triggering the ATR-SOG1-mediated DDR. Upon activation, SOG1 represses cell cycle progression in leaves and prevents extensive meristematic cell death in the radicle, thereby preserving mitotic competency. SOG1 is also essential for activating DDR in the apical root (AR) to inhibit its overgrowth. Additionally, unknown components downstream of ATR contribute to the survival of zdp ape2 embryos. (mC, DNA methylation; Pol, polymerase; Lig1, DNA ligase 1. The starburst indicates an oxidized base.)
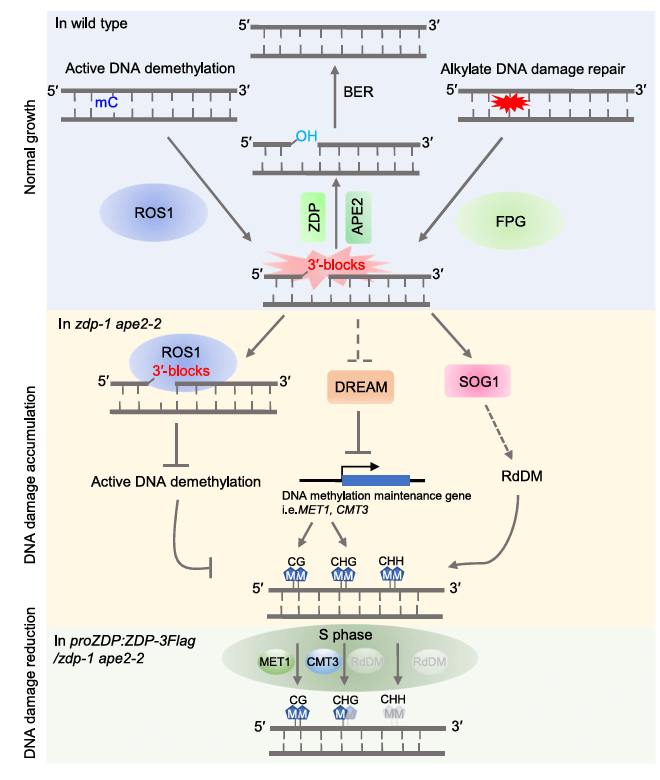
A Model Illustrating How DNA Damage Drives DNA Methylation Establishment and Evolution.
Under normal growth conditions, ROS1-mediated active DNA demethylation and FPG-mediated alkylation DNA damage repair generate DNA 3′-blocks. These blocks are further processed by ZDP and APE2 and subsequently fully repaired by the base excision repair (BER) machinery. In zdp-1 ape2-2 mutants, the accumulation of DNA 3′-blocks perturbs the DREAM complex, leading to hypermethylation in CG and CHG contexts. Additionally, the absence of ZDP/APE2 impairs the processing and turnover of DNA demethylases during active demethylation, potentially causing the persistence of methylation patterns established by these mechanisms. Meanwhile, the disruption of these pathways also activates the RNA-directed DNA methylation (RdDM) pathway, inducing hypermethylation in the CHH context. When DNA damage is reduced (e.g., in the ZDP complementation line), symmetric DNA methylation (CG and CHG) is inherited during the S phase, while asymmetric DNA methylation (CHH) is lost.
